WRN was originally identified as a gene responsible for Werner Syndrome (WS; “Progeria of Adults”). The WRN gene consists of 35 exons that encode a protein of 1,432 amino acids (Yu et al., 1998). The WRN protein contains RecQ-type helicase domains in the central region (Gray et al., 1997) and exonuclease domains in the N-terminal region (Huang et al., 1998). Their preferred substrates resemble various DNA metabolic intermediates, substrates for which helicase and exonuclease activities are thought to function in a coordinated manner. The nuclear localization signal is present at the C-terminal region (Suzuki et al., 2001). Two consensus regions, RecQ helicase conserved region (RQC), the helicase RNaseD C-terminal conserved region (HRDC), are present between the helicase and nuclear localization signal. The biochemical studies, combined with cell biological studies, suggested that this protein is potentially involved in DNA replication, repair, recombination, transcription, telomere maintenance and/or a combination of these events (e.g., repair during replication). The majority of the disease mutations result in truncations of the nuclear localization signals (Friedrich et al., 2010). Precise molecular mechanisms by which mutations in WRN cause the WS phenotype are currently being investigated.
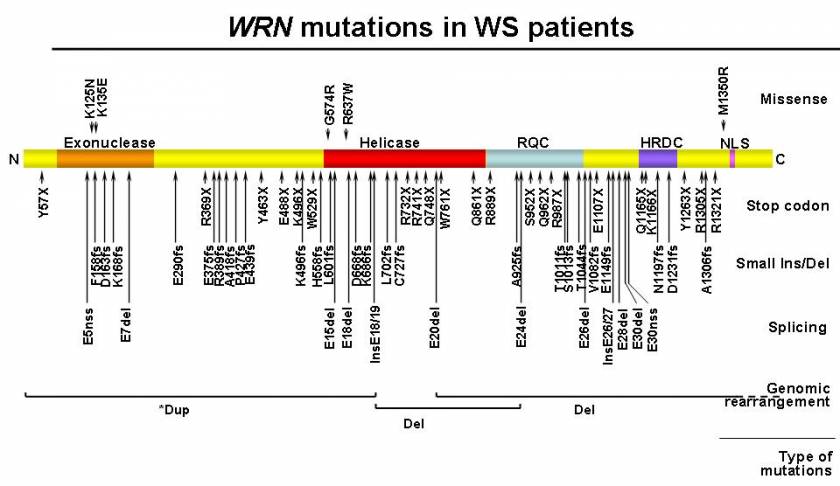
WRN mutations in WS patients. A diagram of the full-length wild type WRN protein is shown, with its N-terminus on the left (N) and C-terminus on the right (C). Known functional domains are marked with darker shades; the exonuclease domain, the helicase domain, the RecQ helicase conserved region (RQC), the helicase RNaseD C-terminal conserved region (HDRC), and the nuclear localization signal (NLS). Mutations are grouped according to canonical classes and further identified by their amino acid changes. Splicing mutations are indicated by the affected exons. Splice mutations that create a new splice site (nss) are indicated as such. Genomic rearrangements, either deletion (Del) or duplication (dup), are shown at corresponding protein locations, with regions extending beyond this figure in dotted lines. * indicates uncertainty in the interpretation of array CGH results (Taken from Friedrich et al., 2010).